Increased Attention on Testing for Oncogenic Drivers in NSCLC Advances the Promise of Precision Medicine
Identification of key oncogenic drivers and the development of targeted therapies with clinical activity in patients harboring actionable mutations have revolutionized the treatment paradigm in non–small cell lung cancer, redirecting attention toward advances in biomarker testing methodologies.
Identification of key oncogenic drivers and the development of targeted therapies with clinical activity in patients harboring actionable mutations have revolutionized the treatment paradigm in non–small cell lung cancer (NSCLC), redirecting attention toward advances in biomarker testing methodologies. This new focus is poised to foster granular refinement of precise, targeted treatment of lung tumors.
Advances in NSCLC research have enabled an understanding of the disease as a collection of molecular subgroups. The proliferation of alteration-matched therapies specific to these subgroups is a prime example of a precision medicine approach. In addition to oncogenic driver mutations, therapeutic response biomarkers have been identified, such as PD-L1 expression as a predictor of immunotherapy efficacy.
Underscoring the importance of biomarker-guided treatment approaches, guidelines for molecular testing in NSCLC include an extensive list of alterations, such as sensitizing EGFR mutations and ALK gene fusions.1,2 The list continues to expand beyond these established canonical markers, with the addition of variants such as MET exon 14 skipping mutations and tumor mutational burden. In fact, the FDA recently approved therapies specific for tumors with these molecular characteristics.3,4
The rapid pace of biomarker discovery, characterization of molecular subtypes of NSCLC, development of matched targeted therapies, and regulatory approval of companion diagnostic tests has accelerated progress in the delivery of optimal care for patients with advanced NSCLC. Areas where continued optimization is particularly emphasized include determining which type of sample(s) should be tested, which biomarkers should be analyzed in different patient subsets, and which assays are most appropriate for specific (sets of) markers, as well as logistical and administrative factors, such as turnaround times and cost/reimbursement considerations.1,5,6
Key Considerations for Molecular Testing in Lung Tumors
As biomarker testing in NSCLC evolves, investigators continue to evaluate testing approaches with the goal of standardizing the process of oncogenic driver identification.
At the 2020 Molecular Analysis for Precision Oncology (MAP) Virtual Congress, held October 9 to 10, 2020, presentations focused on recent developments in molecular testing, including the results of studies comparing testing methods for aberrations in the MET and NTRK genes.
MET Exon 14 Skipping
Gain-of-function alterations in MET, which encodes a receptor tyrosine kinase, drive oncogenesis. One such alteration with important implications for NSCLC is MET exon 14 skipping, resulting from several types of mutation in either exon 14 itself, the adjacent introns, or the flanking splice sites. Regardless, the effect is the same: a critical ubiquitination site is lost, which leads to MET protein accumulation and activation, enhancing MET pathway signal transduction and culminating in tumorigenesis. Previously, immunohistochemistry was typically performed to detect MET overexpression due to copy number changes. However, next-generation sequencing (NGS) is now the preferred testing method because it can also identify MET exon 14 skipping mutations, the primary driver of oncogenesis.7
MET exon 14 skipping mutations are actionable because the resultant protein is responsive to MET inhibition using tyrosine kinase inhibitors (TKIs) such as capmatinib (Tabrecta),7 which was approved in May 2020 for use in adult patients with metastatic NSCLC harboring a MET exon 14 skipping mutation.3 Compared with their sensitivity to specific TKIs, this subset of patients has exhibited lower rates of response to immunotherapy despite frequent tumor expression of PD-L1,8 suggesting a dual predictive role for the MET exon 14 skipping mutation as a biomarker.
In a study presented at MAP 2020, tumor samples from patients with NSCLC and no other driver mutations were tested for MET exon 14 skipping mutations over a period of 14 months. The investigators compared 2 DNA-based methods: NGS on the Ion Proton platform using AmpliSeq technology and fragment analysis using polymerase chain reaction (PCR) and size-based electrophoretic separation of the amplicons for detection of large deletions.9
Of the 87 patient samples tested, 13 were determined to have a MET exon 14 skipping alteration, with 5 harboring splice variants and 8 carrying deletions affecting the splice site. Two of these deletions were large, spanning 41 and 66 base pairs; they were detected by fragment analysis but not NGS. Although NGS is widely considered superior to single-gene assays, these data indicate that it may have limitations in detecting specific alterations and that “complementary methods or large-coverage intron screening could be an alternative” for optimal detection of MET alterations to inform selection of first-line treatment, according to lead study author Romain Loyaux, from the Molecular Oncology Department of Georges-Pompidou European Hospital–APHP in Paris, France.
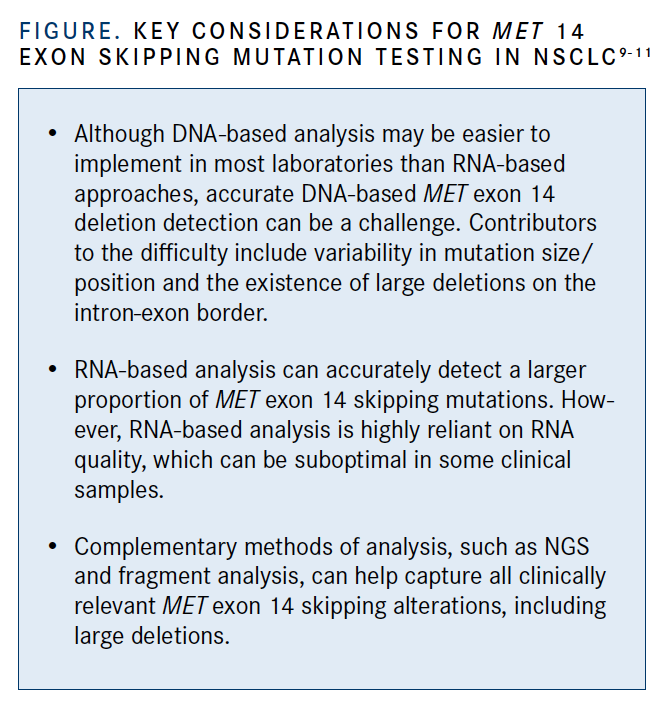
Commenting on the failure of the NGS assay to detect large MET exon 14 deletions, Loyaux stated that “fragment analysis is a cheap and robust method to detect large deletions, especially when no RNA is available” (FIGURE).9-11 He noted that anchored, multiplex, targeted RNA-based NGS, like the technology developed by the Archer company, may be a good alternative when RNA is available; however, it has a 20% failure rate.12
NTRK Fusion Detection
Fusions involving the NTRK genes, which encode a family of receptor tyrosine kinases, result in a constitutively activated chimeric protein that promotes oncogenesis and therefore, can be therapeutically targeted with TKIs.7 Two FDA-approved TKIs, entrectinib (Rozlytrek) and larotrectinib (Vitrakvi), have activity in NTRK fusion–positive solid tumors.13,14
Broad, hybrid-capture DNA-based NGS, with RNA-based anchored multiplex PCR as an adjunct, are currently the primary methods of detecting NTRK gene fusions in patients with lung cancer.7 The availability of entrectinib and larotrectinib will likely foster further development of NTRK fusion detection methods for use in routine clinical practice.
A recent study presented at MAP 2020 evaluated the analytical performance—namely, sensitivity, specificity, and precision—of 3 commonly available RNA-based NGS assays. The assays examined were TruSight Oncology 500 (TSO500) by Illumina, Oncomine Focus Assay (OFA) by Thermo Fisher Scientific, and Fusion- Plex Lung (AFL) by Archer.15
The limits of sensitivity and precision were assessed using droplet digital PCR with admixtures of both NTRK fusion–positive and –negative samples, whereas specificity was evaluated using NTRK fusion–negative clinical samples. The data showed that all 3 NGS assays successfully detected NTRK fusions; however, technical differences between the assays may impact their performance. For instance, although all tested assays demonstrated strong specificity, the sample metrics were variable. Quality control (QC) success rates for OFA and TSO500 were 83% and 77%, respectively, but only 43% of samples on AFL passed all assay QC metrics. Notably, the different assays missed specific NTRK fusions; OFA failed to detect NTRK1-LMNA, NTRK1-TFG, and NTRK2- PAN3, and TSO500 failed to report NTRK3-ETV6 (E5N14) and NTRK3-ETV6 (E5N15).15
Clinical Utility of NGS Panels of Different Sizes
It has been established that multigene panels are superior to single-gene assays for biomarker testing in NSCLC2; however, data to inform clinicians’ selection of specific NGS gene panels have been lacking.
In a recent study presented at MAP 2020, a literature review was conducted to compare 2 commercially available DNA-based NGS gene panels: the Ion AmpliSeq Cancer Hotspot Panel, covering hotspots in 50 genes (Panel 50); and the FoundationOne panel, covering the complete exons of 315 genes (Panel 315). The clinical utility of each panel was determined based on the number of detectable actionable alterations in various solid tumor types that it contained. The data showed a substantial gain in actionability using the larger gene panel, which matched more actionable genetic mutations to FDA-approved or experimental drugs; the number of actionable alterations in various solid tumor types using Panel 315 was a median 50% higher compared with Panel 50 (t test, P <.001). The authors attributed this gain to the inclusion of more genes related to homologous recombination repair deficiency and microsatellite instability/immunotherapy response in the larger panel.16
Looking Ahead
In the current era of precision medicine in lung cancer, defined histological subtyping, oncogenic driver testing, and analysis of tumor PD-L1 expression/immunotherapy sensitivity are crucial steps prior to therapeutic decision-making in NSCLC. As additional targeted agents are investigated in clinical trials and the incidence of their molecular targets are characterized in patient populations, expanded molecular testing may become necessary.
Molecular pathologists will continue to play an integral role in the continuum of care in NSCLC, from diagnosis to clinical decision-making based on biomarker detection. Molecular testing is likely to expand rapidly, and additional molecular subtypes will be identified that help match more patients with the optimal targeted therapies, providing highly personalized treatment plans.
Details of analytical procedures and assays will continue to be refined.17 By combining defined sets of biomarkers with appropriate protocols for collecting NSCLC samples and optimized methods for assessing specific changes, clinicians will be able to actualize the promise of precision medicine for patients with this challenging malignancy.
References:
1. Lindeman NI, Cagle PT, Aisner DL, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med. 2018;142(3):321-346. doi:10.5858/arpa.2017-0388-CP
2. NCCN. Clinical Practice Guidelines in Oncology. Non-small cell lung cancer, version 8.2020. Accessed October 26, 2020. https://bit.ly/2TKomAj
3. FDA grants accelerated approval to capmatinib for metastatic nonsmall cell lung cancer. FDA. May 6, 2020. Accessed November 2, 2020. https://bit.ly/360o7Xg
4. FDA approves pembrolizumab for adults and children with TMB-H solid tumors. FDA. June 16, 2020. Accessed November 2, 2020. https://bit.ly/2HWB64q
5. Smeltzer MP, Wynes MW, Lantuejoul S, et al. The International Association for the Study of Lung Cancer global survey on molecular testing in lung cancer. J Thorac Oncol. 2020;15(9):1434-1448. doi:10.1016/j.jtho.2020.05.002
6. Wempe MM, Stewart MD, Glass D, et al. A national assessment of diagnostic test use for patients with advanced NSCLC and factors influencing physician decision-making. Am Health Drug Benefits. 2020;13(3):110-119
7. Sabari JK, Santini F, Bergagnini I, Lai WV, Arbour KC, Drilon A. Changing the therapeutic landscape in non-small cell lung cancers: the evolution of comprehensive molecular profiling improves access to therapy. Curr Oncol Rep. 2017;19(4):24. doi:10.1007/s11912-017-0587-4
8. Sabari JK, Montecalvo J, Chen R, et al. PD-L1 expression and response to immunotherapy in patients with MET exon 14-altered non-small cell lung cancers (NSCLC). J Clin Oncol. 2017;35(suppl 15):8512. doi:10.1200/JCO.2017.35.15_suppl.8512
9. Loyaux R, Blons H, Garinet S, Urban P, Leger C, Bastide M. MET exon 14 screening strategy: how not to miss large deletions. Ann Oncol. 2020;31(suppl 5): S1217-S1239. doi:10.1016/j.annonc.2020.08.2163
10. Pruis MA, Geurts-Giele WRR, von der TJH, et al. Highly accurate DNAbased detection and treatment results of MET exon 14 skipping mutations in lung cancer. Lung Cancer. 2020;140:46-54. doi:10.1016/j.lungcan.2019.11.010
11. Davies KD, Lomboy A, Lawrence CA, et al. DNA-based versus RNAbased detection of MET exon 14 skipping events in lung cancer. J Thorac Oncol. 2019;14(4):737-741. doi:10.1016/j.jtho.2018.12.020
12. Cohen D, Hondelink LM, Solleveld-Westerink N, et al. Optimizing mutation and fusion detection in NSCLC by sequential DNA and RNA sequencing. J Thorac Oncol. 2020;15(6):1000-1014. doi:10.1016/j.jtho.2020.01.019
13. FDA approves entrectinib for NTRK solid tumors and ROS-1 NSCLC. FDA. Published August 15, 2019. Accessed October 28, 2020. https://bit.ly/3mPhUEB
14. FDA approves larotrectinib for solid tumors with NTRK gene fusions. FDA. Published November 26, 2018. Accessed October 28, 2020. https://bit.ly/381dXZe
15. Bormann Chung C, Lee J, Barritault M, et al. Evaluating targeted next-generation sequencing (NGS) assays and reference materials for NTRK fusion detection. Ann Oncol. 2020;31(suppl 5):S1221. doi:10.1016/j.annonc.2020.08.2172
16. Özdemir B, Charrier M, Gerard CL, et al. Comparison of the clinical utility of two different size next generation sequencing (NGS) gene panels for solid tumours. Ann Oncol. 2020;31(suppl 5):S1219. doi:10.1016/j.annonc.2020.08.2166
17. Pennell NA, Arcila ME, Gandara DR, West H. Biomarker testing for patients with advanced non-small cell lung cancer: real-world issues and tough choices. Am Soc Clin Oncol Educ Book. 2019;39:531-542. doi:10.1200/EDBK_237863