Improved Mutation Profiles Inch Prostate Cancer Algorithms to the Next Level
Advances in molecular profiling have driven development and use of personalized medicine approaches in oncology, enabling detection of biomarkers for predicting prognosis and treatment response and determining inheritable cancer risk.
James Mohler, MD

James Mohler, MD
Knowledge about germline and somatic sequencing is evolving rapidly and helps to inform treatment decisions. With genomic technologies gaining prominence, including next-generation sequencing, the promise of personalized medicine will soon become a reality.
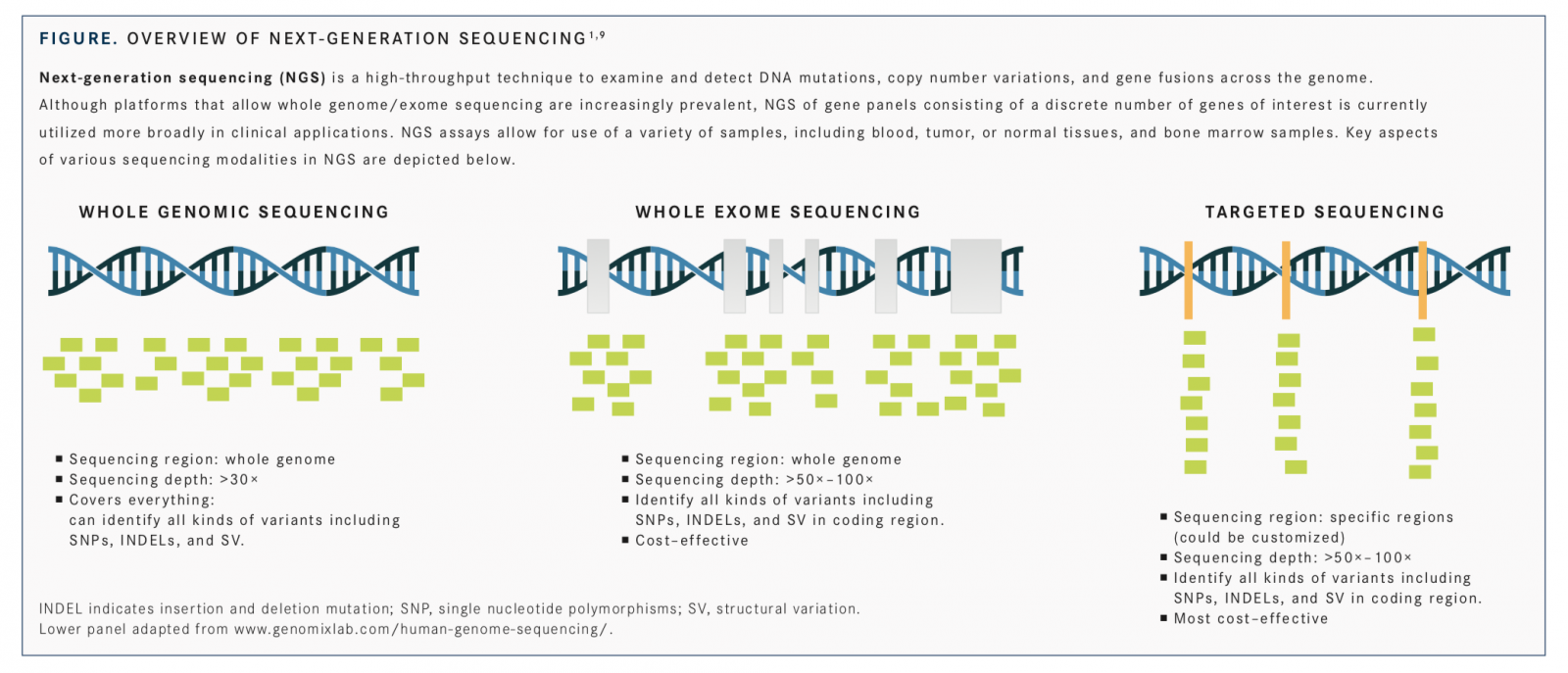
Advances in molecular profiling have driven development and use of personalized medicine approaches in oncology, enabling detection of biomarkers for predicting prognosis and treatment response and determining inheritable cancer risk.1Molecular testing for detecting mutations (deletions, insertions, point mutations), expression changes, amplifications, and gene fusionsof specific biomarkers has been incorporated into recommendations for clinical management of several cancers, including lung,2breast,3,4and colorectal,5,6and is an integral and critical component of personalized oncology practice.1,7,8The veritable explosion of genomic technologies, including next-generation sequencing (NGS) platforms (FIGURE),1,9and their cost decline, along with developments in their regulatory approval by the FDA and acceptance by the Centers for Medicare & Medicaid Services (CMS), are all essential in converting the promise of personalized medicine into reality.1,10,11
The role of germline/somatic mutations as disease drivers in prostate cancer has only recently been appreciated, with the discovery of germline defects in DNA damage repair (DDR) pathway genes, despite long-standing acknowledgment of the contribution of inherited genetic risk in prostate cancer pathogenesis.12,13Compelling evidence for association of germline variants in DDR and mismatch repair (MMR) pathway genes in increased incidence, aggressiveness, and/or response to targeted therapies, especially, has garnered interest and raised significant questions.12,14,15Including recommendations for genetic counseling/testing for DDR/MMR pathway genes in the recent updates to the National Comprehensive Cancer Center (NCCN) Clinical Practice Guidelines for prostate cancer16is a response in integrating molecular testing into clinical practice.
“The knowledge base about the role of genomic and somatic sequencing in guiding both early detection [of] prostate cancer and treatment for prostate cancer is, to put it mildly, evolving rapidly,” James Mohler, MD, associate director for translational research and a professor of oncology at Roswell Park Comprehensive Cancer Center in Buffalo, New York, toldTargeted Therapies in Oncology(TTO) in an interview.
Mohler, who has chaired the NCCN Prostate Cancer Treatment Panel since 2005 and is a member of the panel that drafted the 2019 NCCN guidelines, alluded to the challenge of improving the guidance regarding molecular testing in prostate cancer, given the paucity of data pertaining to demonstrated “clinical utility of knowing the germline and somatic sequencing results.” Mohler also referred to a recent report aimed at providing additional clarification and summarizing the rationale for the guideline updates on molecular testing.16“[This report] attempts to summarize our rationale for the updated recommendations,” he said.
NCCN Guideline Updates for Molecular Testing
The updated NCCN guidelines recommend that clinicians inquire about family and personal history of cancer at the time of initial prostate cancer diagnosis.16In addition, germline genetic testing, with or without pretest genetic counseling, is recommended for patients with prostate cancer and any of the following: a positive family history of cancer (eg, prostate, breast), Ashkenazi Jewish ancestry, or intraductal histology or high-risk, or high-risk, regional, or metastatic prostate cancer, regardless of family history.14,16
The following genes should be included in the germline testing using NGS:MLH1,MSH2,MSH6, andPMS2(associated with Lynch syndrome) andBRCA2,BRCA1,ATM,PALB2, andCHEK2(homologous recombination [HR] genes associated with the DDR pathway). The minimum set of genes in the NGS panel recommended should also include those mentioned above andPALB2,MLH1,MSH2,MSH6, andPMS2.14,16,17Additional genes, such asHOXB13, which is a prostate cancer risk gene18,19but currently has no clear therapeutic implications, may also be considered for testing in certain clinical contexts.
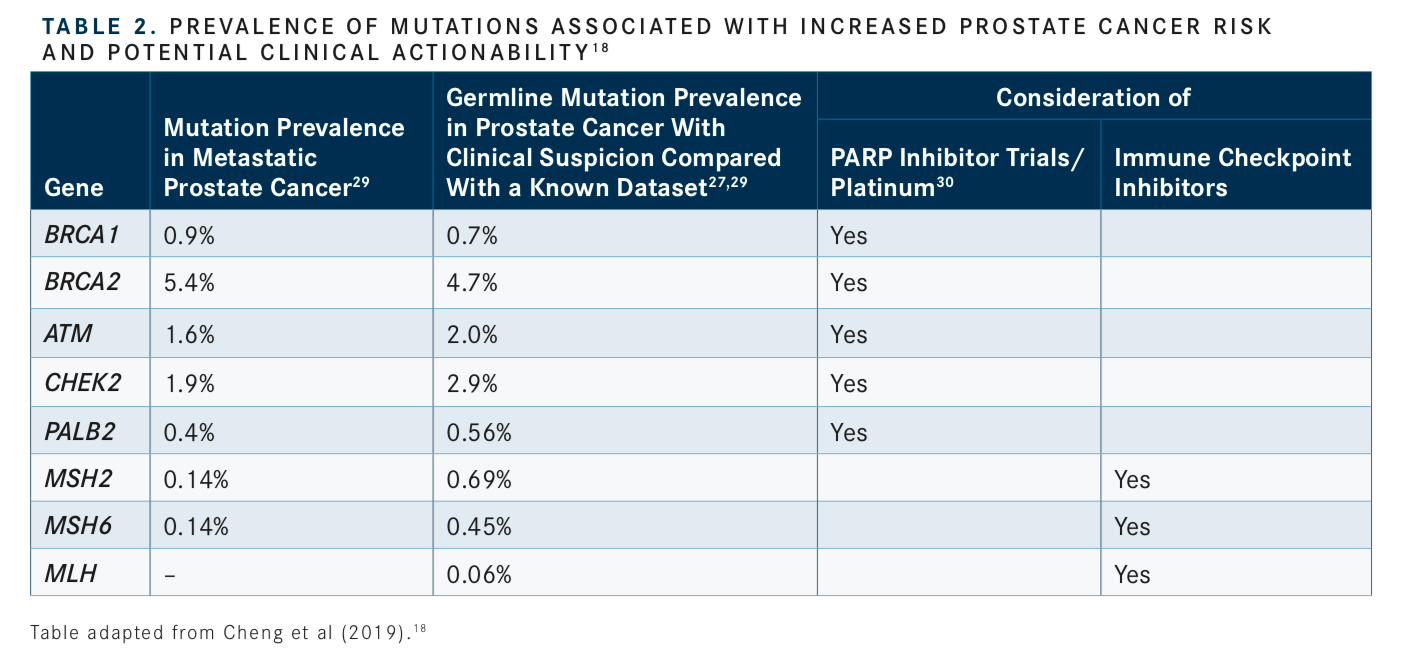
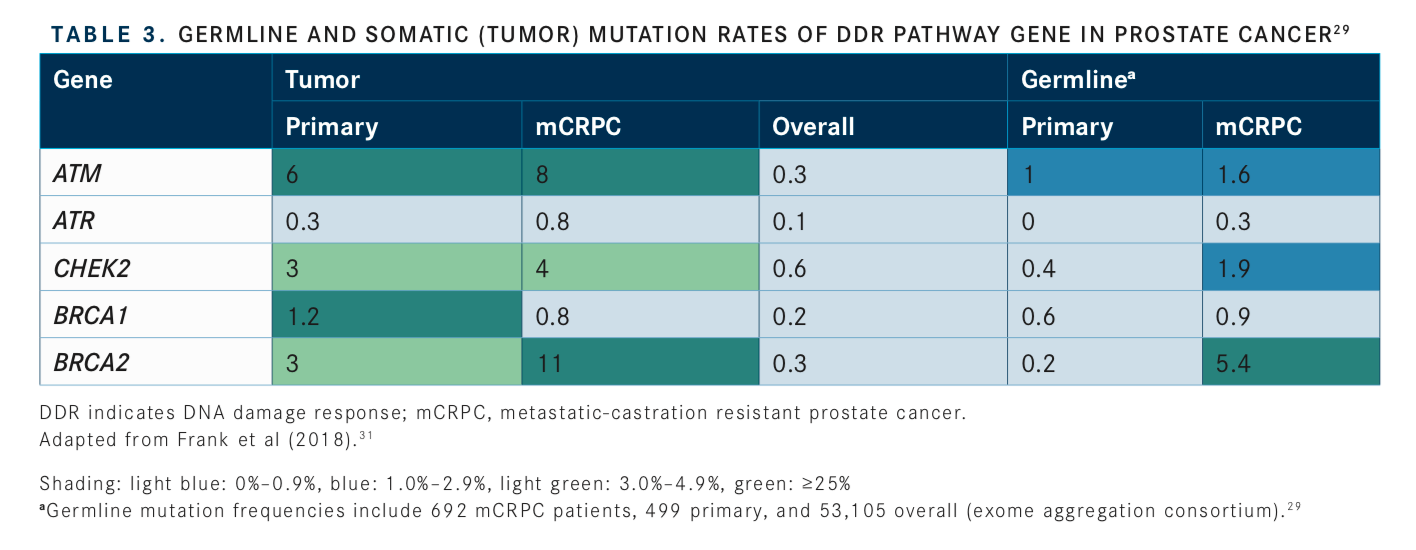
Consideration of somatic (tumor) testing based on risk groups is also recommended for patients with regional or metastatic prostate cancer. If HR gene mutations are detected, referral for genetic counseling to assess for the possibility of hereditary breast and ovarian cancer syndrome is recommended. In patients positive for MSI-H/dMMR mutations, genetic counseling to assess the possibility of Lynch syndrome and, in patients with metastatic castration-resistant prostate cancer (mCRPC), eligibility for pembrolizumab (Keytruda) in second and subsequent lines of treatment should be considered. In patients with low and favorable intermediate-risk prostate cancer and life expectancy ≥10 years, multigene testing can be considered. The rationale for these updates and the current data regarding prevalence of genetic mutations and their association with potential clinical actionability is summarized inTABLES 1to3.14-17, 20-31
The DDR/HR pathway includes PARP enzymes 1 and 2 in addition toBRCA1/2,ATM, and other gene products; in the context of loss of function mutations in genes in the DDR pathway, inhibiting PARP evokes synthetic lethality.32Moreover, the DDR pathway intersects with androgen receptor signalingandrogen-deprivation therapy promotes “BRCAness” in prostate cancer cells and increases their sensitivity to PARP inhibition, which may have therapeutic implications for using androgen signaling inhibitors prior to PARP inhibitor therapy in prostate cancer.21,33,34
Molecular Tests: What, When, Where, and How
“We have been doing this for a long time in lung, breast, and other cancers. I think it is great that prostate cancer is catching up in terms of finding targeted therapies to help treat our patients,” Channing J. Paller, MD, an associate professor of oncology and urology at the Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins Medicine in Baltimore, Maryland, toldTTO, speaking to the significance of the NCCN guideline updates. “The NCCN guidelines are critical in helping insurance pay for standardized testing of patients in terms of both germline and somatic mutations that may guide future treatments and clinical trial eligibility.”In the context of prostate cancer, several molecular tests for evaluating prognostic and predictive serum/urine/tissue-based biomarkers have gained FDA or Clinical Laboratory Improvement Amendments (CLIA) approval.35For instance, Prolaris and the Oncotype DX Genomic Prostate Score test, which assess mRNA expression of multigene panels, may aid in risk stratifying men with low-risk or favorable intermediate-risk disease and deciding between active surveillance and treatment.36,37Decipher Prostate, a test that assesses mRNA expression of 22 genes involved in cell proliferation, migration, tumor motility, androgen signaling, and immune system evasion, may predict metastasis or mortality risk following local therapy.36,37
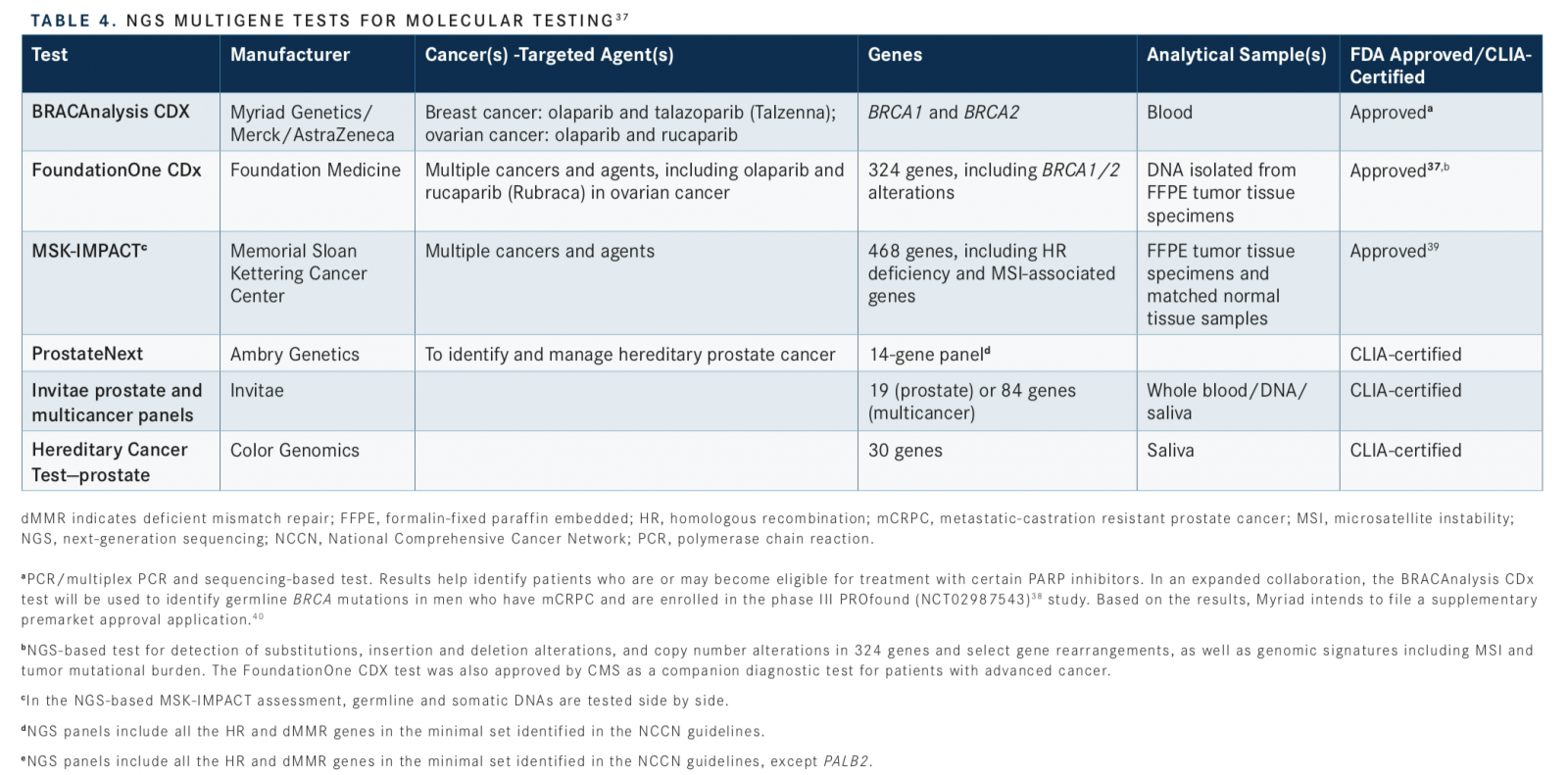
Although commercial NGS assays, which often target exome sequencing, are available, only some have gained FDA approval, as companion diagnostics for multiple molecular-targeted therapies.38Of these, BRACAnalysis CDx, FoundationOne CDx, and MSK-IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets) include HR genes. NGS tests are also available through other companies and CLIA-certified diagnostic laboratories, including ProstateNext (Ambry Genetics), Invitae prostate and multicancer panels (Invitae Genetics), and Hereditary Cancer Testprostate (Color Genomics) (TABLE 437-40).
Germline genetic testing, which can be performed using lymphocyte DNA from blood or a combination of lymphocyte and buccal cells from saliva, can provide information regarding pathogenic variants associated with prostate cancer risk.12,13,17Somatic genetic testing can provide information on clinically actionable alterations that may affect treatment choice or referral to a clinical trial.12,13,17
Although the NCCN updates recommend germline testing for every patient with metastatic prostate cancer and high-risk, localized nonmetastatic disease, somatic testing should be considered for all patients with metastatic disease, including lymph node metastases.16Somatic testing may uncover mutations that are of germline origin, such as founderBRCAmutations, which may change over time genetic instability or therapy-associated selective pressure. This may necessitate germline testing if mutations associated with inherited cancer risk are detected in somatic testing and repeat testing of somatic mutations over the disease course.17,41Given the implications of germline testing data on family members beyond the patient, tumor testing should not be in lieu of germline testing when inherited risk is suspected.17
Questions remain regarding the sequencing of germline/somatic testing, the appropriate tests, and the optimal sample for assessment. Speaking to the context of germline testing, Mohler said that for patients aged <60 years who have an aggressive prostate cancer (Gleason Score 8-10 or Grade Group 4-5) or patients with metastatic prostate cancer (higher mortality risk), the risk of harboring an unfavorable genome may be high enough to warrant germ-line counseling and testing. The data from such testing may have significant implications for first-degree relatives.
“There are many commercially available tests and testing centers currently open, and which ones can be useful in a diagnostic approach [are] yet to be determined,” Paller said. She added that a variety of marketing techniques and clinical utility assessments are used by different testing programs. For example, Invitae espouses an approach to screen 10,000 men with prostate cancer for germ-line mutations, on the heels of a large study that identified positive disease-causing genetic variant mutations in 17% of over 3600 men with a personal history of prostate cancer.27
Addressing the timing of the germline and somatic testing, Paller noted, “[It may] depend on where an individual patient is in their disease course and in the treatment landscape option. For instance, if biopsy specimens uncover clinically actionable somatic mutations, that may drive treatment choice. For patients with biochemically recurrent disease, it is feasible to test only germline samples for mutations, as that is the tissue that may be available.” She alluded to NCT03047135, a clinical trial that is recruiting men with non- metastatic biochemically recurrent prostate cancer and seeks to evaluate germ line testing guided identification of patients with HR gene mutations eligible for PARP inhibitor therapy.
Placing Molecular Testing in the Treatment Algorithm
Integrating molecular testing into the treatment algorithms for prostate cancer is in its fledgling stage but likely to expand soon. “Right now, we only have data of associations of specific mutations with future treatment response. We need additional comparative response data from prospective clinical trials where patients found to have actionable mutations are randomized to targeted therapies, such as PARP inhibitors or checkpoint inhibitors, or standard-of-care treatments. It is going to be very important to continue to acquire more information about response, based upon somatic and genomic sequence data,” Mohler said, speaking to the current state of molecular testing in driving personalized treatment choices.
Studies that can provide supporting data for these include PROfound42 (mCRPC;BRCA1/2andATMmutated; olaparib [Lynparza]), NCT0248440442 (advanced/recurrent tumors, including prostate cancer; with or without somatic or germline DDR mutations; olaparib and durvalumab [Imfinzi]), BRCAAway43(mCRPC; germline or somatic HR pathway mutation-positive; olaparib alone compared with abiraterone [Zytiga] alone and olaparibabiraterone combination), TRITON344(mCRPC;BRCA1/2andATMmutated; rucaparib [Rubraca] monotherapy compared with physicians’ choice of enzalutamide [Xtandi], abiraterone, or docetaxel), Galahad45(mCRPC; DDR mutation positive; niraparib [Zejula]), TRIUMPH46(metastatic hormone-sensitive prostate cancer; DDR mutation positive; rucaparib), and SMMART47(advanced cancers; actionable mutations; targeted therapies).12,13
Looking Ahead: Clinical and Nonclinical Practical Questions
Recent reports have also formulated frameworks for integrating genetic testing into prostate cancer clinical practice.17,18,48Presence of tumor and/or germline mutations in HR-associated genes can help assess eligibility for early use of platinum-based chemotherapy or enrollment in PARP inhibitor clinical trials.17,49,50Initial clinical data indicate that HR deficiency due toBRCAmutation, especiallyBRCA2, predicts response to PARP inhibitors, with emerging evidence suggesting potential susceptibility to this class of agents in the background of mutations in other DDR genes, such asATM, albeit to a lesser extent.15,21Functional HR assays may complement the genetic testing data when determining potential response to PARP inhibitor therapy.21Retrospective analyses suggest thatBRCA2-mutated prostate cancer may be sensitive to platinum-based chemotherapy, and additional data from ongoing studies can help resolve the role of this therapy in DDR mutation positive prostate cancer.15Identification of tumor MSI-H or dMMR, using immunohistochemistry or NGS methods that detect loss of function ofMLH1,MSH2,MSH6, orPMS2,can help identify patients eligible for potential pembrolizumab therapy. Data pertaining to clinical use of pembrolizumab in dMMR/MSI-H prostate cancer is currently limited; for instance, just 2 prostate cancer cases were included in the tumor response data that formed the basis for FDA approval of pembrolizumab in tissue/ site-agnostic dMMR/MSI-H tumors.50,51The opening salvo in the debate on the role of molecular genetic testing in prostate cancer has been launched, marked by the critical NCCN 2019 guideline updates and emerging algorithms to help clarify clinical decision making. With the anticipated data from key clinical studies, a fuller picture of the testing protocols and methodologies, patient criteria for testing use, and application of targeted therapies based on molecular testing data is imminent.
“We are on the cusp of a transformation of treating our patients with prostate cancer differently depending on their mutation profile and status,” Paller said.
In addition to clinical questions, there are barriers and concerns related to other aspects of delivering appropriate genetic testing across the patient population. These include the health, economic/insurance coverage, and emotional consequences for family members of patients in whom molecular testing identifies inherited risk-associated mutations, cost and reimbursement issues pertaining to NGS/genetic testing, uncertainty regarding healthcare policies that may affect the economic burden of molecular testing on patients and the healthcare system, and the national shortage of genetic counselors.12,36
The “financial toxicity” of the potential implications of recommendations for molecular testing in clinical practice guidelines and the impact of uncertainty regarding healthcare policy is particularly challenging and concerning, Mohler noted. “People are going to have to wait for an evolution of guidelines where you get expert opinion driving these choices that must be made on a day-to-day basis in men who are, unfortunately, diagnosed with aggressive prostate cancer,” he added.
Declining testing costs and CMS national coverage determinationapproval of NGS tests as companion diagnostics in advanced cancer have moved the ball forward. However, given the complexity and challenge of resolving clinical and nonclinical issues, Mohler said, “I think it is just as wrong to get sequencing data on everybody diagnosed with prostate cancer as it is wrong to get genomic sequencing on nobody diagnosed with prostate cancer.”
Paller also alluded to the challenges and barriers: “I think if we can overcome some of these concerns and demonstrate clinically meaningful activity, efficacy, and improvement of quality of life with these [actionable mutation-targeted] agents, personalized prostate cancer treatments can become the new standard of care. We are working every day to find more targeted therapy treatments, besides androgen receptor directed therapy, to treat prostate cancer.”
References
- El-Deiry WS, Goldberg RM, Lenz H-J, et al. The current state of molecular testing in the treatment of patients with solid tumors, 2019. CA CancerJ Clin. 2019;69(4):305-343. doi: 10.3322/caac.21560.
- Ettinger DS, Wood DE, Aisner DL, et al. Non-small cell lung cancer, version 5.2017, NCCN Clinical Practice Guidelines in Oncology.J Natl Compr Canc Netw. 2017;15(4):504-535.
- Goetz MP, Gradishar WJ, Anderson BO, et al. NCCN guidelines insights: breast cancer, version 3.2018.J Natl Compr Canc Netw. 2019;17(2):118-126. doi: 10.6004/jnccn.2019.0009.
- Daly MB, Pilarski R, Berry M, et al. NCCN guidelines insights: genetic/familial high-risk assessment: breast and ovarian, version 2.2017.J NatlCompr Canc Netw. 2017;15(1):9-20.
- Benson AB 3rd, Venook AP, Cederquist L, et al. Colon cancer, version 1.2017, NCCN clinical practice guidelines in oncology.J Natl Compr CancNetw. 2017;15(3):370-398.
- Benson AB, Venook AP, Al-Hawary MM, et al. Rectal cancer, version 2.2018, NCCN Clinical Practice Guidelines in Oncology.J Natl Compr CancNetw. 2018;16(7):874-901. doi: 10.6004/jnccn.2018.0061.
- Kamel HFM, Al-Amodi HSAB. Exploitation of gene expression and cancer biomarkers in paving the path to era of personalized medicine. Genomics Proteomics Bioinformatics. 2017;15(4):220-235. doi: 10.1016/j.gpb.2016.11.005.
- Colombo I, Kurnit KC, Westin SN, Oza AM. Moving from mutation to actionability.Am Soc Clin Oncol Educ Book. 2018;38:495-503. doi: 10.1200/EDBK_199665.
- Kamps R, Brandão RD, Bosch BJ, et al. Next-generation sequencing in oncology: genetic diagnosis, risk prediction and cancer classification.Int J Mol Sci. 2017;18(2). doi: 10.3390/ijms18020308.
- Shabani Azim F, Houri H, Ghalavand Z, Nikmanesh B. Next generation sequencing in clinical oncology: applications, challenges and promises: a review article.Iran J Public Health. 2018;47(10):1453-1457.
- Phillips KA. Evolving payer coverage policies on genomic sequencing tests: beginning of the end or end of the beginning?JAMA.2018;319(23):2379-2380. doi: 10.1001/jama.2018.4863.
- Carlo MI, Giri VN, Paller CJ, et al. Evolving intersection between inherited cancer genetics and therapeutic clinical trials in prostate cancer: a white paper from The Germline Genetics Working Group of the Prostate Cancer Clinical Trials Consortium.JCO Precis Oncol. 2018;2018. doi: 10.1200/PO.18.00060.
- Paller CJ1, Antonarakis ES1, Beer TM, et al; PCCTC Germline Genetics Working Group. Germline genetic testing in advanced prostate cancer; practices and barriers: survey results from the Germline Genetics Working Group of the Prostate Cancer Clinical Trials Consortium.Clin Genitourin Cancer. 2019;17(4):275-282.e1. doi: 10.1016/j.clgc.2019.04.013.
- Mohler JL, Antonarakis ES. NCCN guidelines updates: management of prostate cancer.J Natl Compr Canc Netw. 2019;17(5.5):583-586. doi: 10.6004/jnccn.2019.5011.
- Nombela P, Lozano R, Aytes A, Mateo J, Olmos D, Castro E. BRCA2 and other DDR genes in prostate cancer.Cancers (Basel). 2019;11(3):pii:E352. doi: 10.3390/cancers11030352.
- Mohler JL, Antonarakis ES, Armstrong AJ, et al. Prostate cancer, version 2.2019, NCCN Clinical Practice Guidelines in Oncology.J Natl Compr Canc Netw. 2019;17(5):479-505. doi: 10.6004/jnccn.2019.0023.
- Cheng HH, Sokolova AO, Schaeffer EM, Small EJ, Higano CS. Germline and somatic mutations in prostate cancer for the clinician.J Natl Compr Canc Netw. 2019;17(5):515-521. doi: 10.6004/jnccn.2019.7307.
- Giri VN, Knudsen KE, Kelly WK, et al. Role of genetic testing for inherited prostate cancer risk: Philadelphia Prostate Cancer Consensus Conference 2017.J Clin Oncol. 2018;36(4):414-424. doi: 10.1200/JCO.2017.74.1173.
- Cooney KA, Beebe-Dimmer JL. HOXB13 mutations and prostate cancer risk.BJU Int. 2016;118(4):496-497. doi: 10.1111/bju.13477.
- Lavery A, Gilson C2, Chowdhury S. PARP inhibitors and stratified treatment of prostate cancer.Expert Rev Anticancer Ther. 2016;16(12):1213-1215. doi: 10.1080/14737140.2016.1243474.
- Virtanen V, Paunu K, Ahlskog JK, Varnai R, Sipeky C, Sundvall M. PARP inhibitors in prostate cancerthe preclinical rationale and current clinical development.Genes (Basel). 2019;10(8):pii:E565. doi: 10.3390/genes10080565.
- Isaacsson Velho P, Silberstein JL, Markowski MC, et al. Intraductal/ductal histology and lymphovascular invasion are associated with germline DNA-repair gene mutations in prostate cancer.Prostate. 2018;78(5):401-407. doi: 10.1002/pros.23484.
- Taylor RA, Fraser M, Livingstone J, et al. Germline BRCA2 mutations drive prostate cancers with distinct evolutionary trajectories.Nat Commun. 2017;8:13671. doi: 10.1038/ncomms13671.
- Risbridger GP, Taylor RA, Clouston D, et al. Patient-derived xenografts reveal that intraductal carcinoma of the prostate is a prominent pathology in BRCA2 mutation carriers with prostate cancer and correlates with poor prognosis.Eur Urol. 2015;67(3):496-503. doi: 10.1016/j.eururo.2014.08.007.
- Latham A, Srinivasan P, Kemel Y, et al. Microsatellite instability is associated with the presence of Lynch syndrome pan-cancer.J Clin Oncol. 2019;37(4):286-295. doi: 10.1200/JCO.18.00283.
- Abida W, Cheng ML, Armenia J, et al. Analysis of the prevalence of microsatellite instability in prostate cancer and response to immune checkpoint blockade.JAMA Oncol. 2019;5(4):471-478. doi: 10.1001/jamaoncol.2018.5801.
- Nicolosi P, Ledet E, Yang S, et al. Prevalence of germline variants in prostate cancer and implications for current genetic testing guidelines.JAMA Oncol. 2019;5(4):523-528. doi: 10.1001/jamaoncol.2018.6760.
- Lynparza (olaparib) granted breakthrough therapy designation by US FDA for treatment of BRCA1/2 or ATM gene mutated metastatic-castration resistant prostate cancer [news release]. Cambridge, UK: AstraZeneca; January 28, 2016.astrazeneca.com/media-centre/press-releases/2016/Lynparza-Olaparib-granted-Breakthrough-Therapy-Designation-by-US-FDA-for-treatment-of-BRCA1-2-or-ATM-gene-mutated-metastatic-Castration-Resistant-Prostate-Cancer-28012016.html. Accessed August 12, 2019.
- Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer.N Engl J Med. 2016;375(5):443-453. doi: 10.1056/NEJMoa1603144.
- Wyatt AW, Annala M, Aggarwal R, et al. Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer.J Natl Cancer Inst. 2017;109(12):1-9. doi: 10.1093/jnci/djx118.
- Frank S, Nelson P, Vasioukhin V. Recent advances in prostate cancer research: large-scale genomic analyses reveal novel driver mutations and DNA repair defects.F1000Res.2018;7:pii:F1000 Faculty Rev-1173. doi: 10.12688/f1000research.14499.1.
- Lord CJ, Tutt AN, Ashworth A. Synthetic lethality and cancer therapy: lessons learned from the development of PARP inhibitors.Annu Rev Med.2015;66:455-470. doi: 10.1146/annurev-med-050913-022545.
- Asim M, Tarish F, Zecchini HI, et al. Synthetic lethality between androgen receptor signalling and the PARP pathway in prostate cancer.Nat Commun.2017;8(1):374. doi: 10.1038/s41467-017-00393-y.
- Li L, Karanika S, Yang G, et al. Androgen receptor inhibitor-induced “BRCAness” and PARP inhibition are synthetically lethal for castration-resistant prostate cancer.Sci Signal. 2017;10(480):pii:eaam7479. doi: 10.1126/scisignal.aam7479.
- Kohaar I, Petrovics G, Srivastava S. A rich array of prostate cancer molecular biomarkers: opportunities and challenges.Int J Mol Sci. 2019;20(8):pii:E1813. doi: 10.3390/ijms20081813.
- Bernard B, Flaig TW. Point-prostate cancer genomic analysis: routine or research only?Oncology (Williston Park). 2018;32(12):607-609.
- Nagahashi M, Shimada Y, Ichikawa H, et al. Next generation sequencing‐based gene panel tests for the management of solid tumors.Cancer Sci. 2019;110(1):6-15. doi: 10.1111/cas.13837.
- Allegretti M, Fabi A, Buglioni S, et al. Tearing down the walls: FDA approves next generation sequencing (NGS) assays for actionable cancer genomic aberrations.J Exp Clin Cancer Res. 2018;37(1):47. doi: 10.1186/s13046-018-0702-x.
- De Bono JS, Hussain M, Thiery-Vuillemin A, et al. PROfound: a randomized phase III trial evaluating olaparib in patients with metastatic castration-resistant prostate cancer and a deleterious homologous recombination DNA repair aberration.J Clin Oncol. 2017;35(suppl 15; abstr TPS5091). doi: 10.1200/JCO.2017.35.15_suppl.TPS5091.
- Myriad will seek FDA approval of BRACAnalysis CDx® as a companion diagnostic for Lynparza (olaparib) in men with metastatic castrate-resistant prostate cancer [press release].Salt Lake City, UT: Myriad Genetics Inc; August 7, 2019. investor.myriad.com/news-releases/news-release-details/myriad-will-seek-fda-approval-bracanalysis-cdxr-companion. Accessed August 13, 2019.
- Ternyila D. Integrating genetic testing into prostate cancer guidelines leads to improved treatment. Targeted Oncology™ website. targetedonc.com/publications/targeted-therapy-news/2019/may-2019/integrating-genetic-testing-into-prostate-cancer-guidelines-leads-to-improved-treatment-decisions. Published May 17, 2019. Accessed August 9, 2019.
- Karzai F, VanderWeele D, Madan RA, et al. Activity of durvalumab plus olaparib in metastatic castration-resistant prostate cancer in men with and without DNA damage repair mutations.J Immunother Cancer.2018;6(1):141. doi: 10.1186/s40425-018-0463-2.
- Reichert Z, Carneiro BA, Daignault-Newton S, et al. A randomized phase II trial of abiraterone, olaparib or abiraterone + olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair defects.J Clin Oncol. 2017;35(suppl 15; abstr TPS5086). doi:10.1200/JCO.2017.35.15_suppl.TPS5086.
- Ryan CJ, Watkins SP, Despain D, et al. Trial of rucaparib in prostate indications 3 (TRITON3): an international, multicenter, randomized, open-label phase 3 study of rucaparib vs physician’s choice of therapy for patients (Pts) with metastatic castration-resistant prostate cancer (mCRPC) associated with homologous recombination deficiency (HRD).J Clin Oncol. 2017;35(suppl 15; abstr TPS5087). doi: 10.1200/JCO.2017.35.15_suppl.TPS5087.
- Smith MR, Sandhu SK, Kelly WK, et al. Phase II study of niraparib in patients with metastatic castration-resistant prostate cancer (mCRPC) and biallelic DNA-repair gene defects (DRD): preliminary results of GALAHAD.J Clin Oncol. 2019;37(suppl 7; abstr 202). doi: 10.1200/JCO.2019.37.7_suppl.202.
- Markowski MC, Wang H, Sullivan R, et al. Phase II trial of rucaparib (without ADT) in patients with metastatic hormone-sensitive prostate cancer harboring germline DNA repair gene mutations (TRIUMPH).J Clin Oncol. 2018;36(suppl 15; abstr TPS5095). doi: 10.1200/JCO.2018.36.15_suppl.TPS5095.
- Mitri ZI, Parmar S, Johnson B, et al. Implementing a comprehensive translational oncology platform: from molecular testing to actionability.J Transl Med. 2018;16(1):358. doi: 10.1186/s12967-018-1733-y.
- Cheng H, Powers J, Schaffer K, Sartor O. Practical methods for integrating genetic testing into clinical practice for advanced prostate cancer.Am Soc Clin Oncol Book. 2018;(38):372-381. doi: 10.1200/EDBK_205441.
- Warner EW, Yip SM, Chi KN, Wyatt AW. DNA repair defects in prostate cancer: impact for screening, prognostication and treatment.BJU Int. 2019;123(5):769-776. doi: 10.1111/bju.14576.
- Marcus L, Lemery SJ, Keegan P, Pazdur R. FDA approval summary: pembrolizumab for the treatment of microsatellite instability-high solid tumors.Clin Cancer Res.2019;25(13):3753-3758. doi: 10.1158/1078-0432.CCR-18-4070.
- Broderick JM. FDA approves pembrolizumab for microsatellite instability-high and mismatch repair deficient cancers. OncLive® website. onclive.com/web-exclusives/fda-approves-pembrolizumab-for-microsatellite-instabilityhigh-and-mismatch-repair-deficient-cancers. Published May 23, 2017. Accessed August 22, 2019.
