Advancements in Targeting NRG1 Fusions in Solid Tumors
Gene fusions aretumor-specific hybrid genes formed by structural chromosomal rearrangements; these changes lead to expression of chimeric proteins that may cause dysregulation of cellular functions. Gene fusions are involved in key pathogenic mechanisms (eg, gene truncations mediating inactivation of tumor suppressor genes) that result in cancer development and progression.1
The discovery of tumor-specific gene fusions has led to advances in precision cancer therapy, such as development of targeted therapies that specifically bind to and inhibit the activity of oncogenic driver proteins.1 The tyrosine kinase inhibitor imatinib blocks the oncogenic kinase activity of BCR-ABL, a chimeric protein that is encoded by the BCR-ABL1 gene. Imatinib became the first gene fusion–targeted therapy when it was approved by the FDA in 2001 to treat chronic myeloid leukemia and in 2006 to treat acute lymphoblastic leukemia.2 In 2011, crizotinib was approved by the FDA to treat patients with non–small cell lung cancer (NSCLC) that harbors ALK fusions.3 These breakthroughs have paved the way for the development of other innovative personalized therapies that target oncogenic driver proteins encoded by rare gene fusions.
In 2014, chromosomal gene copy number analysis and lung adenocarcinoma (ADC) sample transcriptome sequencing in patients who had never smoked tobacco led to the detection of CD74–NRG1, a somatic gene fusion. These fusions were found mostly in tumor samples from women with invasive mucinous adenocarcinoma (IMA).4
Results of a retrospective analysis of more than 21,000 tumor samples confirmed the presence of neuregulin 1 (NRG1) fusions in various types of malignant tumors, including NSCLC, breast cancer, cholangiocarcinoma, ovarian cancer, pancreatic cancer, renal cell carcinoma (RCC), bladder cancer, and sarcoma.5 Although NRG1 fusions are rare, their presence in multiple types of cancer make them targets of novel treatments that are in development for difficult-to-treat cancers.
Neuregulin-1
Neuregulins (NRGs), also known as heregulins, are proteins that serve as activating ligands for cell-surface receptors of the HER (ERBB) family of receptor tyrosine kinases and are involved in cellular differentiation and proliferation. Dysregulation of these processes can lead to the development of cancer.6
NRG1 proteins are encoded by the NRG1 gene. NRG1 binds to HER3 and HER4 receptors found on the surface of many types of cells within the epithelial, mesenchymal, and neuronal lineages.5
NRG1 transcription produces 6 different forms of NRG1 that include at least 31 different isoforms. All NRG1 isoforms include an extracellular epidermal growth factor (EGF)-like domain that binds to and triggers the activation of HER receptor tyrosine kinases. Unlike most NRG1 isoforms that undergo proteolytic cleavage leading to the release of the EGF-like domain, type 3 NRG1s have a membrane-tethered EGF-like domain that mediates signaling via interaction between cells.6
HER3 Activation
The HER family of cell-surface receptors triggers intracellular signaling pathways when activated by cognate ligands vital to healthy cellular function and activity.7,8 Overactivation of HER3 has been implicated in oncogenesis and resistance to medical cancer treatments.6
Dimerization is the process by which 2 receptors pair to trigger kinase activity and initiate intracellular signaling; homodimerization involves a pairing of the same type of receptor, whereas heterodimerization involves the pairing of 2 different receptors.9-11 Both HER2 and HER3 rely upon heterodimerization to become active; the former has no known activating ligand, and the latter lacks intrinsic kinase activity.7,8
NRG1 can activate HER receptors via autocrine or paracrine signaling.9-11 Upon binding with NRG1, HER3 undergoes a conformational change that exposes its dimerization domain and facilitates pairing with its preferred partner, HER2. Because HER3 contains multiple sites for the p85 regulatory subunit of PI3K, HER2-HER3 dimerization amplifies PI3K/AKT signaling and can also trigger other signaling pathways, such as MAPK.7,8
However, NRG1 fusions can create NRG1 fusion proteins that cause pathologic activation of HER3 receptors and lead to tumorigenesis, the process of unregulated cell growth and survival that creates tumor cells.4,6,12
Findings from in vitro preclinical studies revealed that the presence of CD74-NRG1 leads to enhanced cancer cell proliferation and tumor growth. In H1568 lung cancer cells, the ectopic expression of CD74-NRG1 resulted in enhanced colony formation in soft agar assays.4
Malignant transformation driven by HER3 mutations was also observed in vitro in a preclinical model of colon cancer.4 Moreover, the heterodimerization of HER2 and HER3 is among the most malignant of known mitogenic interactions. This could be attributed to several molecular events, including the slow endocytosis of HER2-containing dimers and rapid recycling to the cell surface. Additionally, ligand-mediated activation of HER3 results in tyrosine phosphorylation that can recruit PI3K without the adapter protein GRB2, allowing for the evasion of ligand-dependent degradation of HER3. Unregulated HER2/HER3 dimerization activation of the PI3K and MAK pathways can induce cellular proliferation and suppress apoptosis.6
NRG1 Fusions as Driver Alterations and Therapeutic Targets
Tumor cells often carry numerous genomic alterations; some are associated with oncogenic activity, whereas others do not promote cancer growth. Driver alterations impair cell division and checkpoints, leading to abnormal cell proliferation and tumorigenesis. Passenger mutations may not have any effect on cells but are present in tumors as a consequence of cell division that is carried along the clonal expansion of cancerous cells.13
When a new driver alteration in cancer is identified, researchers and drug developers can begin to formulate novel therapeutics that specifically target the oncogenic activity of that driver alteration to inhibit tumor growth and survival.Other common oncogenic drivers (ie, mutated KRAS, EGFR, ALK, ROS1, and RET) are frequently absent in tumors that have NRG1. Consequently, NRG1 fusions are considered unique and actionable oncogenic driver alterations.14
IMA accounts for 2% to 10% of all lung ADC and is typically associated with KRAS mutations; however, tumors that harbor NRG1 fusions test negative for mutated KRAS and other common mutations, indicating that NRG1 fusions are the driver alteration for these cases of IMA. Therefore, NRG1 fusion–targeted therapies could fill a treatment gap for some patients with cancers that are unresponsive to drugs that were developed for other oncogenic targets.4
Incidence of NRG1 Fusions
The presence of NRG1 fusions in tumor cells is rare, but advancements in technologies that can detect fusions may reveal higher incidences and identify new fusion partners. The incidence of NRG1 fusions in solid tumors was investigated in a retrospective study by Jonna et al. The researchers surveyed a deidentified molecular profiling database for solid tumors that underwent fusion testing; all cases submitted to a Clinical Laboratory Improvement Amendments-certified laboratory for comprehensive genomic profiling from September 2015 to December 2018 were included. All unique cases that underwent successful4 fusion testing for targeted RNA sequencing were identified, and a board-certified pathologist reviewed all histologic characteristics.5
A total of 21,858 tumor specimens from unique patients across a variety of cancer types were successfully evaluated; rates of NRG1 fusions found in samples of solid tumors are shown in Table 1.5 The overall incidence of NRG1 fusions was 0.2%. Among the cancers in which NRG1 fusions were the most prevalent in tumors were cholangiocarcinoma (0.5%), pancreatic ductal ADC (PDAC; 0.5%), (0.5%), RCC (0.5%), and ovarian cancer (0.4%). These results suggested that NRG1 fusions represent a potential therapeutic target for a variety of cancer types.5
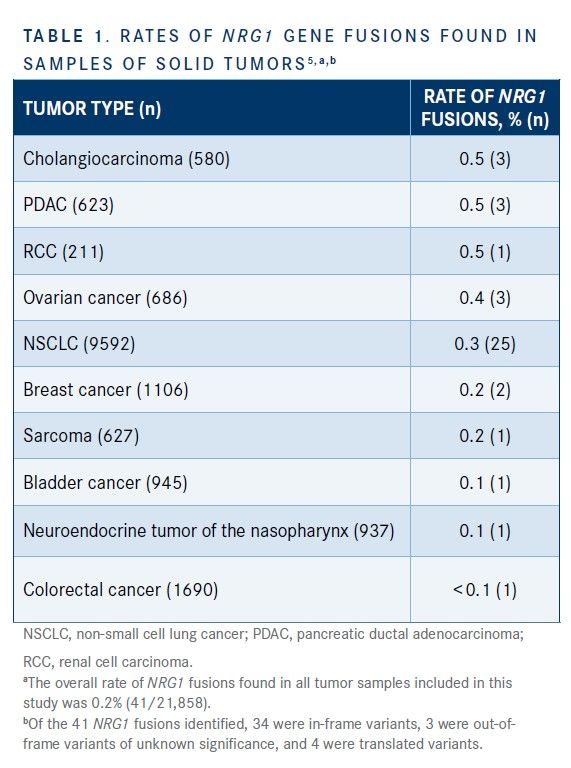
Drilon et al presented large-scale genomic profiling data from patients with NSCLC to support a basket trial (also known as a bucket trial or tumor-agnostic trial) approach to drug development for NRG1 rearrangements that extended beyond NSCLC to other cancers.14
In this study, 8984 solid tumor datasets from The Cancer Genome Atlas and 17,985 solid tumor datasets detected using the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) assay were analyzed for the presence of NRG1 fusions and fusion partners in multiple cancer types (ie, breast, head and neck, kidney, lung, ovarian, pancreatic, prostate, and uterine cancers).14
A total of 10 NRG1 fusions from both datasets were identified: 7 in lung ADC, 2 in PDAC, and 1 in breast carcinoma. In the MSK-IMPACT dataset alone, NRG1 rearrangements were detected in 3 patients with lung ADC, 1 patient with PDAC, and 1 patient with HER2-positive breast cancer. These results supported the authors’ assertion that basket trials of NRG1 fusion–targeted therapies would be useful in drug development. Furthermore, if cancers respond to NRG1 fusion–targeted therapies regardless of their histology, tumor-agnostic drug development could be adopted to investigate the use of HER2/HER3 inhibitors.14
Identified NRG1 Fusion Partners
As noted by Jonna et al in their aforementioned article, NRG1 has numerous and diverse fusion partners within and across cancer types (Table 2).5 In this study, NSCLC tumor samples had the highest number of NRG1 fusion partners detected; CD74 was the most common, followed by SDC4, SLC3A2, TNC, MDK, ATP1B1, DIP2B, RBPMS, MRPL13, ROCK1, DPYSL2, and PARP8. Three NRG1 fusion partners were identified in ovarian cancer (SETD4, TSHZ2, and ZMYM2) and pancreatic cancer (ATP1B1, CDH1, and VTCN1).5
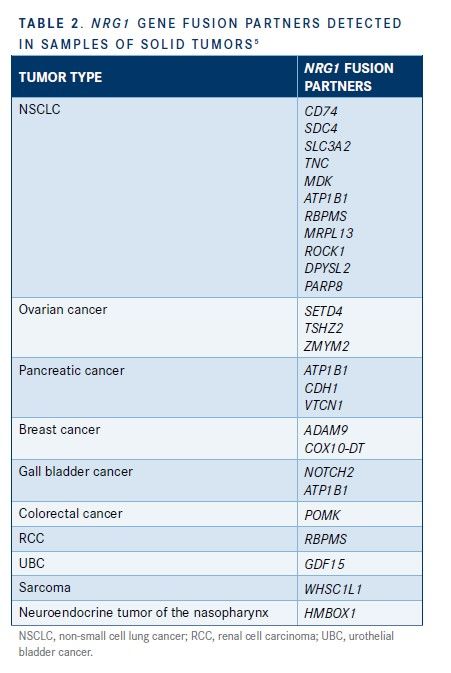
This study also discovered the co-occurrence of NRG1 fusions with other common oncogenic mutations.5NRG1 fusions are mutually exclusive with oncogenic driver mutations of EGFR, KRAS, ALK, ROS, and RET. In patients with tumors that harbor NRG1 fusions, using treatments that target this gene fusion with agents that are directed at activity from other actionable mutations may be useful.5,15
Detection of Novel NRG1 Fusions and Identification of Partners
Jonna et al sought to characterize NRG1 fusion events in solid tumors and identify new fusion partners. Tumor samples (N = 44,570) were submitted for clinical molecular profiling using RNA-based next-generation sequencing (NGS) and were retrospectively analyzed for NRG1 fusion events. All NRG1 fusions that had at least 3 junction reads were identified for manual review and for characterization of fusion class, intact functional domains, domain prediction, breakpoints, frame retention, and co-occurring alterations by NGS.16
Fusion events were identified in 82 tumor samples (0.2%). Tumor types involved were NSCLC (54.0%), breast cancer (11.0%), ovarian cancer (7.0%), pancreatic cancer (7.0%), cholangiocarcinoma (6.0%), colorectal cancer (5.0%), and other malignancies(10.0%). Among the fusion events detected, 42 unique partners were identified; CD74 (23.0%), ATP1B1 (9.0%), SLC3A2 (7.0%), RBPMS (6.0%), and SDC4 (4.0%) were the most common.16
Approximately half of the fusion events identified were expected to contain a transmembrane domain provided by the fusion partner. Lung and pancreatic cancers had the highest rates of inclusion of transmembrane domains; in other types of cancer, the fusion partners did not include a transmembrane domain. In 15% of cases, the chimeric transcript was expected to cause increased expression of NRG1.16
Genomic Testing for NRG1 Fusions
The feasibility of using genomic testing of rare cancers to identify personalized treatment strategies was investigated in a study by Kee et al.17 Half of all cases analyzed had at least 1 actionable mutation that could guide treatment decisions. Approved therapeutic options are available for alterations in EGFR, BRAF, HER2, KRAS, MET, ROS1, PI3K, and PDGFR, as well as in gene fusions involving ALK, NTRK, RET,and ROS1, among others.2 Accurate and rapid detection of chromosomal rearrangements can help improve understanding of cancer pathogenesis, precision of diagnoses, and treatment personalization.10
Genomic testing is the only way to confirm whether a tumor has an NRG1 fusion. Many types of tests for detecting gene fusions are available, but some are more sensitive than others.18 Fluorescence in situ hybridization (FISH) and reverse-transcription polymerase chain reaction (RT-PCR) have been the most used techniques to detect gene mutations, including fusions. However, because of the diversity and complexity of NRG1 fusion partners and the large intronic regions that characterize NRG1, these traditional DNA-based methods have frequently failed to detect NRG1 fusions. Now considered the gold standard for detecting NRG1 fusions, RNA-based NGS has a higher sensitivity for genetic rearrangements that increases its ability to detect these gene fusions.8
According to data from a multicenter registry study, RNA-based methods were the most commonly used technique to detect NRG1 fusions (79.5%), followed by FISH (12.0%) and DNA-based methods (9.4%).8 The use of RNA-based NGS resulted in identification of more than 10,000 distinct fusions in solid tumors.19
RNA-Based NGS
NRG1 fusion proteins cause pathologic activation of HER3 signaling in tumors harboring NRG1 fusions, leading to tumor growth and survival.2,5,11 To create these proteins, cells with NRG1 fusions convert and shorten the information in DNA to RNA. Therefore, RNA-based NGS may detect proteins actively produced by a cell; this technology can overcome read-length limitations observed with DNA sequencing techniques, resulting in a higher sensitivity for detecting gene fusions.20,21
As part of a global, multicenter registry study that examined the clinical and pathological features of NRG1 fusion–positive NSCLC and clinical outcomes of patients with this disease subtype, Duruisseaux et al used several genomic testing techniques to identify NRG1 fusion–positive tumors. A total of 80 NRG1 fusion–positive tumors were identified; 53 (66.0%) via RNA-based NGS, 14 (18.0%) via DNA-based NGS, and 13 (16.0%) via FISH. The researchers concluded that RNA-based NGS was an important component of NRG1 fusion detection.22
RT-PCR and FISH
RT-PCR is a cyclic DNA-based testing method that was commonly used to detect gene mutations and fusions; however, this technique lacks the ability to detect novel fusion partners, and there are currently no FDA-approved assays available.19
FISH was also a widely used method for detecting gene fusions based on its ability to determine the location of chromosomal breakpoints. This method requires the use 4 of fluorescent probes that bind to specific complementary target sequences. The FISH method does not require living cells for analysis; however, the results are biased towards detection of chromosomal translocations and lacks sensitivity in detecting intrachromosomal events, such as intragenic deletion.19
Therapeutic Challenges in NRG1 Fusion–PositiveCancer
Following the identification of NRG1 fusions as potential driver alterations, Duruisseaux et al launched a study to examine the clinical and pathological features of NRG1 fusion–positiveNSCLC and clinical outcomes of patients with this disease subtype. The primary end point was best response to systemic therapy and progression-free survival (PFS).22
Most of the patients harboring NRG1 fusion–positive tumors were women (64.0%), and many had never smoked (58.0%). Nearly all of these patients (91.0%) had IMA, and none of the tumors presented high PD-L1 expression.22
Of the 12 patients with stage IV disease treated with the HER2 inhibitor afatinib, 11 were evaluable. The best response in this cohort was progressive disease, which occurred in 6 patients (55.0%). Two patients (18.0%) had a partial response, and 2 patients (18.0%) had stable disease. The median PFS among these patients was 3.5 months.22
Nineteen patients received platinum-based chemotherapy.Among this cohort, the best response was stable disease, which was noted in 8 patients (47.0%). Disease progressed in 7 patients (41.0%), and 2 patients (12.0%) had a partial response. None of the patients in this study responded to single-agent PD-L1 therapy or chemoimmunotherapy (ie, carboplatin, pemetrexed, pembrolizumab).22
These results indicated that NRG1 fusion–positive tumors do not respond as well to current standard-of-care treatments as do those that do not harbor the fusion; novel, targeted therapies are needed to improve clinical outcomes of patients presenting with this disease subtype.22
Overall Survival and Disease-Free Survival
The presence of NRG1 fusions has been correlated with poor prognosis. Shin et al reported on the prevalence of NRG1 fusions in a cohort of patients with IMA and investigated oncogenic function, pathways, and clinical implications of their findings. In this study, NRG1 fusion–positive tumors were found in 16 of 59 patients with IMA; 13 of these patients had tumors positive for the NRG1-SLC3A2, and 3 had tumors positive for the NRG1-CD74.23
Patients with NRG1 fusion–positive IMA had a median overall survival of 52 months; OS was not reached in patients with NRG1 fusion–negative IMA. Also, the median disease-free survival was 22 months in patients with NRG1 fusion–positive IMA as compared with 79 months in patients with NRG1 fusion–negative IMA.23
Moreover, the ectopic expression of the SLC3A2-NRG1 in lung cancer cells resulted in greater cell migration, proliferation, and tumor growth in vitro and in xenograft models.23
Conclusions
Advances in genomic technologies have enabled the detection and characterization of rare genomic aberrations such as NRG1 fusions, which have been found in various types of malignant tumors. The characterization of NRG1 fusions as actionable genomic events and their detection in the absence of other common actionable genomic variations offers the unique opportunity to develop and guide personalized treatment of hard-to-treat malignancies. The current lack of standard-of-care therapy for patients with NRG1 fusion–positive cancer highlights the need to identify specific treatments for validation across tumor types in a tumor-agnostic approach.
References
1. Mertens F, Johansson B, Fioretos T, Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15(6):371-381. doi:10.1038/nrc3947
2. Drugs@FDA: FDA-approved drugs. Approval date(s) and history, letters, labels, reviews for NDA 021588. United States Food and Drug Administration. Accessed September 9, 2021. https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event=overview.process&ApplNo=021588
3. Drugs@FDA: FDA-approved drugs. Approval date(s) and history, letters, labels, reviews for NDA 202570. United States Food and Drug Administration. Accessed September 9, 2021. https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event=overview.process&ApplNo=202570
4. Fernandez-Cuesta L, Plenker D, Osada H, et al. CD74-NRG1 fusions in lung adenocarcinoma. Cancer Discov. 2014;4(4):415-422.
doi:10.1158/2159-8290.CD-13-0633
5. Jonna S, Feldman RA, Swensen J, et al. Detection of NRG1 gene fusions in solid tumors. Clin Cancer Res. 2019;25(16):4966-4972.
doi:10.1158/1078-0432.CCR-19-0160
6. Fernandez-Cuesta L, Thomas RK. Molecular pathways: targeting NRG1 fusions in lung cancer. Clin Cancer Res. 2015;21(9):1989-1994. doi:10.1158/1078-0432.CCR-14-0854
7. Jacob W, James I, Hasmann M, Weisser M. Clinical development of HER3-targeting monoclonal antibodies: perils and progress. Cancer Treat Rev. 2018;68:111-123. doi:10.1016/j.ctrv.2018.06.011
8. Mishra R, Patel H, Alanazi S, Yuan L, Garrett JT. HER3 signaling and targeted therapy in cancer. Oncol Rev. 2018;12(1):355. doi:10.4081/oncol.2018.355
9. Russo A, Lopes AR, Scilla K, et al. NTRK and NRG1 gene fusions in advanced non-small cell lung cancer (NSCLC). Precis Cancer Med. 2020;3(14). doi:10.21037/pcm.2020.03.02
10. Laskin J, Liu SV, Tolba K, et al. NRG1 fusion-driven tumors: biology, detection, and the therapeutic role of afatinib and other ErbB-targeting agents. Ann Oncol. 2020;31(12):1693-1703. doi:10.1016/j.annonc.2020.08.2335
11. Nagasaka M, Ou SHI. Neuregulin 1 fusion-positive NSCLC. J Thorac Oncol. 2019;14(8):1354-1359. doi:10.1016/j.jtho.2019.05.015
12. Mota JM, Collier KA, Costa RLB, et al. A comprehensive review of heregulins, HER3, and HER4 as potential therapeutic targets in cancer. Oncotarget. 2017;8(51):89284-89306. doi:10.18632/oncotarget.18467
13. Haber DA, Settleman J. Cancer: drivers and passengers. Nature. 2007;446(7132):145-146. doi:10.1038/446145a
14. Drilon A, Somwar R, Mangatt BP, et al. Response to ERBB3-directed targeted therapy in NRG1-rearranged cancers. Cancer Discov. 2018;8(6):686-695. doi:10.1158/2159-8290.CD-17-1004
15. Heining C, Horak P, Uhrig S, et al. NRG1 fusions in KRAS wild-type pancreatic cancer. Cancer Discov. 2018;8(9):1087-1095. doi:10.1158/2159-8290.CD-18-0036
16. Jonna S, Feldman R, Ou SHI, et al. Characterization of NRG1 gene fusion events in solid tumors. J Clin Oncol. 2020;38(suppl 15):3113. doi:10.1200/JCO.2020.38.15_suppl.3113
17. Kee D, Kondrashova O, Ananda S, et al. NOMINATOR: feasibility of genomic testing of rare cancers to match cancer to treatment. J Clin Oncol. 2020;38(suppl 15):103. doi:10.1200/JCO.2020.38.15_suppl.103
18. Heyer EE, Deveson IW, Wooi D, et al. Diagnosis of fusion genes using targeted RNA sequencing. Nat Commun. 2019;10(1):1388. doi:10.1038/s41467-019-09374-9
19. Schram AM, Chang MT, Jonsson P, Drilon A. Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat Rev Clin Oncol. 2017;14(12):735-748. doi:10.1038/nrclinonc.2017.127
20. Sussman RT, Oran AR, Paolillo C, Liberman D, Morrissette JJD, Rosenbaum JN. Validation of a next-generation sequencing assay targeting RNA for the multiplexed detection of fusion transcripts and oncogenic isoforms. Arch Pathol Lab Med. 2020;144(1):90-98. doi:10.5858/arpa.2018-0441-OA
21. Teixidó C, Giménez-Capitán A, Molina-Vila MA, et al. RNA analysis as a tool to determine clinically relevant gene fusions and splice variants. Arch Pathol Lab Med. 2018;142(4):474-479. doi:10.5858/arpa.2017-0134-RA
22. Duruisseaux M, Liu SV, Han JY, et al. NRG1 fusion-positive lung cancers: clinicopathologic profile and treatment outcomes from a global multicenter registry. J Clin Oncol. 2019;37(suppl 15):9081. doi:10.1200/JCO.2019.37.15_suppl.9081
23. Shin DH, Lee D, Hong DW, et al. Oncogenic function and clinical implications of SLC3A2-NRG1 fusion in invasive mucinous adenocarcinoma of the lung. Oncotarget. 2016;7(43):69450-69465. doi:10.18632/oncotarget.11913
