Three Experts Weigh In on Plasma-Based Molecular Profiling in Oncology
Guardant360 is a highly sensitive digital sequencing system that requires only a 10 mL blood sample, and allows for simultaneous assessment of more than 60 actionable genes in circulating, cell-free DNA.
1Because it is noninvasive, this method may be used to re-assess evolving tumors without a repeat biopsy.
At the American Society of Clinical Oncology (ASCO) 2014 meeting, Morelli et al reported use of this technology to detect, in a single test, both somatic mutations and copy number alterations in patients with treatment-refractory metastatic colorectal cancer (mCRC).2They demonstrated the coexistence of multiple acquired somatic mutations and copy number alterations in patients with epidermal growth factor receptor (EGFR) antibody-refractory mCRC (Figure 1).2
FIGURE 1. Gene copy number alterations detected using the Guardant360 system.2
Gene copy number alterations detected using the Guardant360 system
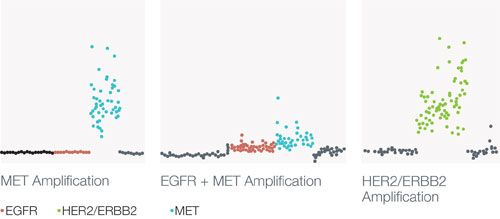
FIGURE 2.
Evolution of genetic changes
3
in a patient with IBC.
FIGURE 2. Evolution of genetic changes in a patient with IBC.
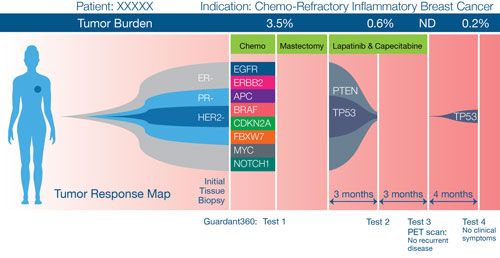
At the 2014 San Antonio Breast Cancer Symposium (SABCS), Austin et al reported genomic alterations over the course of disease in a patient with inflammatory breast cancer (IBC).Figure 2shows the evolution of genomic alterations in this patient, with 15 alterations detected after neoadjuvant therapy, 3 alterations detected after 3 months of adjuvant therapy, and no alterations detected at the end of adjuvant therapy. Notably, 4 months after completing adjuvant therapy, the patient showed a new mutation (before detectable clinical progression).3Targeted Oncology: asked 3 experts in tumor genomics to comment on the clinical potential and application of molecular tests such as Guardant360.
DAVID R. GANDARA, MD,is director of the thoracic oncology program, professor, and senior advisor to the director of the University of California Davis Comprehensive Cancer Center. He is also the founding cochair for the National Cancer Institute (NCI) Investigational Drug Steering Committee, chair for the NCI Lung Correlative Science Committee, and chair of the Southwest Oncology Group (SWOG) Lung Committee.
Q: Why is genomic testing important in lung cancer?GANDARA:Lung cancer is one of the most genomically complex cancers, and sorting out what is actually driving the patient’s cancer, and what is an unrelated abnormality, or a comutation, is challenging. More and more we are into an algorithm, which requires periodic repeat tumor biopsies after every therapy, particularly in these oncogene-driven lung cancers. This is also challenging, because many patients may have a new metastasis in a hard-to-biopsy site (eg, brain, bone), and such biopsies are invasive and costly. Use of cfDNA testing allows you to do repeat testing on a sequential basisevery 2 months if you like—to see if you can detect these new potential drivers and anticipate them ahead of time.
Q: How are these testing platforms (eg, Guardant360) being used in lung cancer clinical practice, and what do you think will be their potential use in the future?GANDARA:About a year and a half ago, we were doing single-gene and multiplex testing in lung cancer, and we had predicted we would be moving into tissue-based NGS. Here it is now, just 16 months later, and [it’s] now routine. Moreover, the next future, plasma-based testing is also here. So right now, the percentage of patients getting this type of testing could be perhaps 1%, and most of that is people doing research, but ask the same question 1 year from nowI think it’s going to be quite different.
Q: Are any large-scale initiatives now using this technology for lung cancers?GANDARA:The SWOG has a large clinical trial about to begin (S1403) in patients with untreated, newly diagnosed, stage 4, EGFR-mutated lung cancer to examine the impact of dual EGFR inhibition. This trial will employ a serial plasma DNA (mutational) analysis, in an effort to see if we can predict early recurrence, and the likelihood of the acquired resistance mechanism.
PHILLIP C. MACK, PHD,is an associate adjunct professor and director of molecular pharmacology in the department of internal medicine and the division of hematology and oncology at the UC Davis Comprehensive Cancer Center.
Q: What are your perspectives on the performance and clinical utility of tests such as Guardant360 in oncology?MACK:This technology can serve as a substitute when a biopsy is not possible. We’re not clear as yet if it will be as routinely predictive as having a piece of tumor tissue, but it certainly can be very informative. Secondly, it can be very useful for following adaptive changes over the course of treatment, for instance, early detection of the emergence of resistance mutations. Also, the ability of this technology to observe tumor DNA copy number abnormalities in plasma is very forward looking.
Q: Can you comment on the performance of this technology relative to tissue biopsy in addressing issues like intratumor heterogeneity?MACK:It’s not clear whether, even in plasma, you will be able to pick up every genetic variation out there, nor would you be able to state definitively that mutation X and mutation Y are occurring in the same tumor cells. However, it is reasonable to speculate that the most aggressive forms of the tumor, even if they represent only a small portion of the overall tumor burdenfor instance, metastatic variants—would be leaking detectable DNA into the peripheral circulation, and therefore would be highly therapeutically informative, possibly more so than a biopsy of the primary lesion.
Q: How might more comprehensive testing across multiple tumor types be beneficial to expand the use of targeted therapies in cancer?MACK:A number of cancer types are at the forefront of targeted, precision therapy, including lung cancer. It is not entirely clear whether this is due to unique properties of those cancers, or just a failure to identify similar oncogenic drivers in other tumor types. So once we analyze all cancer types using these systematic screens, we may identify, at least in small patient subsets, some tumor dependencies that would make themselves amenable to targeted therapies. This will be particularly important for rare tumors that are not as well studied as the major forms.
EDWARD KIM, MD, PHD,is an assistant professor of medicine in the department of internal medicine and the division of hematology and oncology at the UC Davis Comprehensive Cancer Center.
Q: How does biopsy-free genomic profiling using cfDNA offer an advantage over tissue-based analyses?KIM:Tumor biopsies are potentially limited by a sampling bias. We know from studies of multiple tumor types that there can be incredible heterogeneity within a single patient’s tumor. This heterogeneity exists within an individual lesion as well as between different metastatic sites and the primary tumor. Circulating cfDNA may provide a more representative sample of tumor DNA, although this is admittedly based on the assumption that tumor DNA is shed from tumors in a randomly distributed fashion. Tumor biopsies also provide a snapshot in time, and genomic profiling of a tumor prior to treatment will likely be different after multiple lines of treatment. Biopsy-free genomic profiling, on the other hand, can be done with a simple blood test and is therefore significantly more amenable to serial testing to provide up-to-date genomic profiling data, and also makes it possible to obtain information in cases where tissue access is limited due to location or tumor yield (eg, pancreatic cancer).
Q: What are some of the limitations of these types of analyses?KIM:There are many limitations to applying the knowledge from genomic profiling to treatment decisions. Some of these limitations apply to all methods of genomic profiling, regardless of whether it is through direct analysis of tumor tissue or isolation of cell-free circulating DNA. These limitations include the potential for finding many genomic changes that are not actionable, in terms of an available therapy that is either approved or in development. There may also be an available therapy, but not for the specific disease application, thus limiting its true availability. Even if a drug is available, there may not always be a correlation between the identified genomic abnormality and response to the drug.
The greatest utility in genomic profiling is to provide potentially useful information that can guide patients toward clinical trials. In patients with disease refractory to standard treatments (eg, in mCRC), biopsy-free assays provide a mechanism to obtain genomic profiling of a patient’s tumor at the time of progression, as opposed to analyzing the original tumor biopsy or resection specimen, which may no longer be relevant after multiple lines of treatment.
Q: How might biopsy-free genomic profiling using cfDNA be important in guiding treatment for patients with GI cancers (eg, mCRC)?KIM:We are approaching a point where our understanding of tumor biology, targeted therapies, and associated resistance mechanisms, and rapid drug development will be primed for widespread use of genomic profiling. Biopsy-free genomic profiling has the potential to significantly augment the patient population that can be tested.
References
- Talasaz AA, Mortimer S, Schiller B, et al. Comprehensive profiling of cancer in real-time by digital sequencing of cell-free DNA from 520 patients with metastatic solid tumors and its clinical application. Poster presentation at EORTC.
- Morelli MP, Overman M, Vilar-Sanchez E, et al. High frequency of concurrent gene mutations and copy number alterations in circulating cell-free DNA (cfDNA) from refractory metastatic CRC patients. ASCO 2014.
- Austin L, Fortina P, Sebisanovic D, et al. Circulating tumor DNA (ctDNA) provides molecular monitoring for inflammatory breast cancer (IBC). San Antonio Breast Cancer Symposium.